Basic concepts¶
This guide explains Ploomber’s core concepts.
Ploomber allows you to quickly turn a collection of scripts, notebooks, or functions into a data pipeline by following three conventions:
Each task is a function, script or notebook.
Tasks declare their dependencies using an
upstreamvariable.Tasks declare their outputs using a
productvariable.
A simple pipeline¶
Let’s say we want to build a pipeline to plot some data. Instead of coding everything in a single file, we’ll break down logic in three steps, which will make our code more maintainable and easier to test:
Note
A pipeline is also known as a directed acyclic graph (or DAG). We use both of these terms interchangeably.
In a Ploomber pipeline, outputs (also known as products) from one
task become inputs of “downstream” tasks. Hence, “upstream” dependencies read
from left to right. For example, raw is an “upstream” dependency of clean.
An “upstream” dependency implies that a given task uses its upstream products
as inputs. Following the pipeline example, clean uses raw’s products, and plot uses clean’s products.
Ploomber supports three types of tasks:
Python functions (also known as callables)
Python scripts/notebooks (and their R equivalents)
SQL scripts
You can mix any combination of tasks in your pipeline. For example, you can dump data with a SQL query, then plot it with Python.
Defining a pipeline¶
To execute a pipeline, Ploomber needs to know the location of the
task’s source code (source key), and the location of the task’s products
(product key). You can do this via a pipeline.yaml file:
tasks:
# this is a sql script task
- source: raw.sql
product: [schema, name, table]
# ...
# this is a function task
# "my_functions.clean" is equivalent to: from my_functions import clean
- source: my_functions.clean
product: output/clean.csv
# this is a script task (notebooks work the same)
- source: plot.py
product:
# scripts always generate a notebook (more on this in the next section)
nb: output/plots.ipynb
Note
You can set a task name using name. If not present, Ploomber infers it from the source value by removing the extension to the file’s name.
Once you have a pipeline.yaml file, you can run it with:
ploomber build
Ploomber keeps track of source changes to skip up-to-date tasks. If you run
ploomber build again, only tasks whose source code has changed
are executed. This helps iterate faster, as changes to the pipeline only
trigger the least number of tasks.
Tip
You can use the resources_ section in a task definition to tell
Ploomber to track the content of other
files. Click here to learn more.
For a full reference on pipeline.yaml files see: Spec API (pipeline.yaml).
Let’s now see how to use scripts and notebooks as pipeline tasks.
Tasks: scripts/notebooks¶
Jupyter notebooks files (.ipynb) contain both code and output; while convenient, keeping code and outputs in the
same file makes version control (i.e., git) difficult.
Our recommended approach is to use scripts as sources. However, thanks to the
integration with Jupyter, you can open scripts as notebooks. The
following image shows a side-by-side comparison of the same source code
as .py (script) and as a .ipynb (notebook) file:

Note that the .py script has some # %% comments. Such markers allow us
to delimit code cells and render the .py file as a notebook.
Note
The # %% is one way of representing .py as notebooks. Ploomber
uses jupytext to perform the conversion, other formats such as the
“light” (# +) format work too. Editors such as VS Code, Spyder, and
PyCharm support the “percent” format.
To keep the benefits of the .ipynb format, Ploomber creates a
copy of your scripts, converts them to .ipynb at runtime and executes them. This is a
crucial concept: scripts are part of your project’s source code, but executed
notebooks are pipeline products.
Note
Even though we recommend the use of .py files, you can still use regular .ipynb files as sources if you prefer so.
To know more about integration with Jupyter notebooks, see the Jupyter integration guide.
R scripts/notebooks are supported as well.
upstream and product¶
To specify task dependencies, include a special parameters cell in your
script/notebook. Following the example pipeline, clean has raw
as an upstream dependency as the raw task is an input to
the clean task. We establish this relation by declaring an upstream
variable with a list of task names that should execute before the file we’re
editing. If a script/notebook has no dependencies, set upstream = None.
# %% tags=["parameters"]
upstream = ['raw'] # this means: execute raw.py, then clean.py
product = None
Important
product = None is a placeholder. It states that our script takes an input
parameter called product, but the actual value is automatically replaced
at runtime, we explain this in the upcoming section.
Note
the # %% markers only apply to scripts.
Click here
for information on adding tags to .ipynb files.
The cell injection process¶
Note
For tips on troubleshooting pipeline loading, see Troubleshooting pipeline loading.
Let’s review the contents of a sample clean.py file:
import pandas as pd
# %% tags=["parameters"]
upstream = ['raw']
product = None
# %%
df = pd.read_csv(upstream['raw']['data'])
# some data cleaning code...
# %%
# store clean data
df.to_csv(str(product['data']), index=False)
This code will break if we run it: We declared raw as
an upstream dependency, but we don’t know where to load our inputs from, or
where to save our outputs.
When executing your pipeline, Ploomber injects a new cell into each
script/notebooks, with new product and upstream variables that
replace the original ones by extracting information from the
pipeline.yaml:
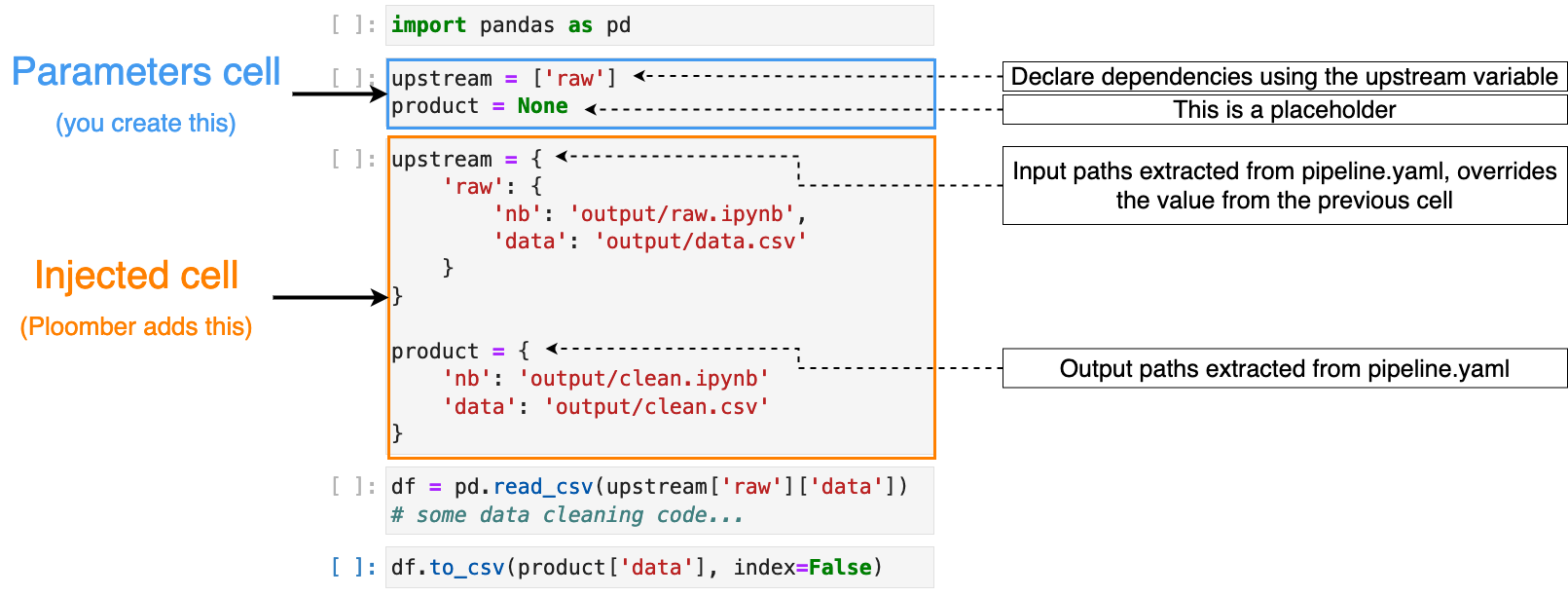
As you can see in the image, the task in the picture has an upstream
dependency called raw. Thus, the injected cell has a dictionary that gives
you the products of raw, which we use as input, and a new product
variable that we use to store our outputs.
The cell injection process also happens when opening the notebook/script in Jupyter. Learn more about Cell injection and Jupyter integration.
Note
When using jupyter notebook, scripts open automatically as
notebooks. If using jupyter lab, you have to right-click and select the
notebook option.
Since scripts/notebooks always create an executed notebook, you must specify where to save such a file. A typical task declaration looks like this:
tasks:
- source: plot.py
# output notebook
product: output/plots.ipynb
If the source script/notebook generates more than one output, create a
dictionary under product:
tasks:
- source: plot.py
product:
# if the script generates other products, use "nb" for the notebok
nb: output/plots.ipynb
# ...and any other keys for other files
data: output/data.csv
Note
The name of keys in the product dictionary can be chosen freely so as to be descriptive of the outputs (e.g. data, data_clean, model, etc.)
To inject cells manually, users can run:
ploomber nb --inject
However, if the same source appears more than once, --inject will pick the first declared task and inject those parameters.
Here is an example where template.ipynb appears in two different tasks
tasks:
- source: template.ipynb
name: task-a
product: output/template-task-a.ipynb
params:
some_param: param-a
- source: template.ipynb
name: task-a-suffix
product: output/template-task-a-suffix.ipynb
params:
some_param: param-a-suffix
- source: template.ipynb
name: task-b-suffix
product: output/template-task-b-suffix.ipynb
params:
some_param: param-b-suffix
By using the inject-priority parameter in setup.cfg, we can specify which set of parameters to inject:
To inject param-a to task-a :
[ploomber]
entry-point = path/to/pipeline.yaml
inject-priority = task-a
Use wildcards to inject multiple parameters (*)
To inject param-a-suffix to task-a-suffix, and param-b-suffix to task-b-suffix :
[ploomber]
entry-point = path/to/pipeline.yaml
inject-priority = *-suffix
This covers scripts and notebooks as tasks, if you want to learn how to use functions as tasks, keep scrolling, otherwise, skip to the end.
It is also possible to use placeholders in the pipeline.yaml file.
For example, the following file uses a placeholder some_param.
# Content of pipeline.yaml
tasks:
- source: print.py
name: print
product:
nb: 'output/{{some_param}}/notebook.html'
papermill_params:
log_output: True
params:
some_param: '{{some_param}}'
This placeholder should be defined inside the env.yaml file, more on that here.
Tasks: functions¶
You can also use functions as tasks, the following section explains how.
upstream and product¶
The only requirement for a function to be a valid task is to have a product parameter.
import pandas as pd
def clean(product):
# save output using the product argument
df.to_csv(product)
Note
If the function generates many products, this becomes a dictionary, for example: product['one'], and product['another'].
If the task has upstream dependencies, add an upstream parameter:
import pandas as pd
def clean(product, upstream):
df_input = pd.read_csv(upstream['task_name'])
df.to_csv(product)
When resolving dependencies, Ploomber will look for references such as
upstream['task_name'], then, during execution, Ploomber will pass the
requested inputs. For example, upstream={'task_name': 'path/to/product/from/upstream.csv'}
This covers scripts and functions as tasks, if you want to learn how to use SQL scripts as tasks, keep scrolling, otherwise, skip to the end.
Tasks: SQL¶
SQL tasks require more setup because you have to configure a client to
connect to the database. We explain the product and upstream mechanism
here; an upcoming guide describes how to configure database clients.
upstream and product¶
SQL scripts require placeholders for product and upstream. A script
that has no upstream dependencies looks like this:
CREATE TABLE {{product}} AS -- {{product}} is a placeholder
SELECT * FROM my_table WHERE my_column > 10
In your pipeline.yaml file, specify product with a list of 3
or 2 elements: [schema, name, table] or [name, table]. If using a
view, use [schema, name, view]. For example:
Say you have product: [schema, name, table] in your pipeline.yaml file.
The {{product}} placeholder is replaced by schema.name:
CREATE TABLE schema.name AS
SELECT * FROM my_table WHERE my_column > 10
If the script has upstream dependencies, use the {{upstream['task_name']}}
placeholder:
CREATE TABLE {{product}} AS
SELECT * FROM {{upstream['task_name']}} WHERE my_column > 10
{{upstream['task_name']}} tells Ploomber to run the task with the name
'task_name' and to replace {{upstream['task_name']}} with the
product of such task.
Clients¶
To establish a connection with a database, you have to configure a client.
All databases that have a Python driver are supported, including systems like
Snowflake or Apache Hive. To learn more, see the SQL guide.
Where to go from here¶
We’ve created many runnable templates to help you get up and running, check out our Downloading templates guide.
If you want to read about advanced features, check out User Guide.
The pipeline.yaml API offers a concise way to declare
pipelines, but if you want complete flexibility, you can use the underlying Python
API, Click here to learn more, or click here to see an example.